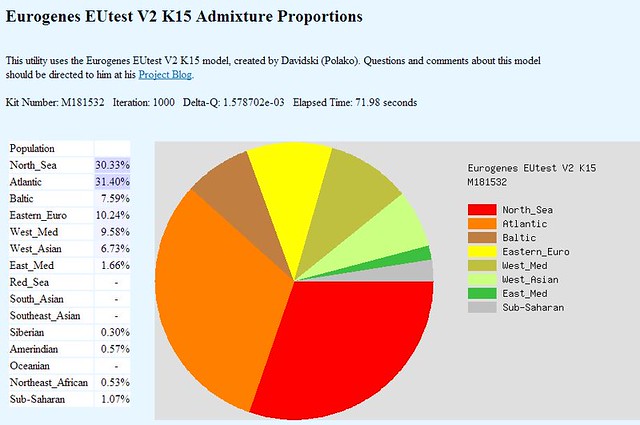
I’m back with a bit more genetic information about our James A. Crider & Elvina Tennessee Thurmond. In my last post I shared some chromosome painting comparisons between myself and my genetic relative, Bev, who is also descended from James and Elvina. There were two matching segments that were fairly obvious to detect, and two that were much more complex. I decided to take the tricky ones and run them through a different utility to see if that would shed more light on the subject. Just as a point of reference, here is how Eurogenes EU15 V2 paints my entire genome:
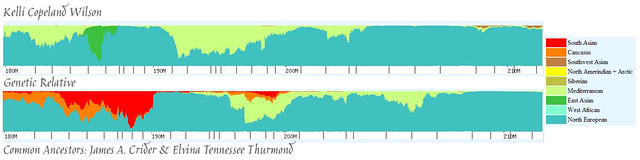
Let’s start with our matching segment on Chromosome 2. Here is the Eurogenes K9 painting:
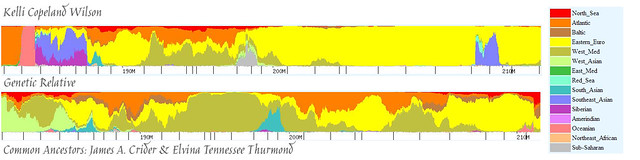
And, here’s what Eurogenes EU15 V2 came up with:
Okay! Now we’re getting somewhere. The Eurogenes EU15 V2 test is the most recent version of the Eurogenes utility and supercedes all previous versions. I still like K9 for its simplicity but, as you can see, the EU15 version gives us a lot more information and makes it easier to identify the places where we match (or don’t). For this particular segment I chose to mark the areas where we differ significantly, as that helps us to focus more closely on where we match.
For example, on the bottom left of my segment you can see an area with Oceanian, blending into a section of Southeast Asian/Siberian/a little strip of Red Sea (Middle East), followed by a spike of South Asian. In the same place on Bev’s chromosome we find West Asian mixed with Western Mediterranean, followed by some South Asian mixed with Oceananian. In the middle of my chromosome we see West Asian mixed with North African, whereas Bev has South Asian and a little Siberian. At the right side of my chromosome we find Southeast Asian mixed with Red Sea, and Bev has Oceanian.
So, what does that leave us with? Based on our knowledge of where we differ (and it is pretty significant in those locations), I feel pretty comfortable saying that we match in all of the red, orange, brown, and yellow areas (and maybe, but less likely, the sort of olive green areas). In other words, the ancestor who gave this DNA to Bev and I was a mix of North Sea, Atlantic, Baltic, and Eastern European (with possibly some Western Mediterranean). In other words, this little section is a nice combination Western European and Eastern European. Given what we know about the Crider family from the paper trails we’ve followed, we know that they were originally from Germany (which is situated right in the middle of Western and Eastern Europe). Combined with the chromosome painting information, I’ll just take a guess and say that this segment belongs to James Crider.
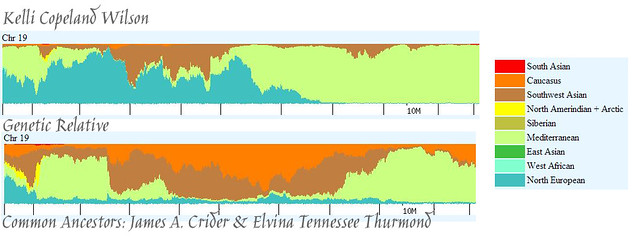
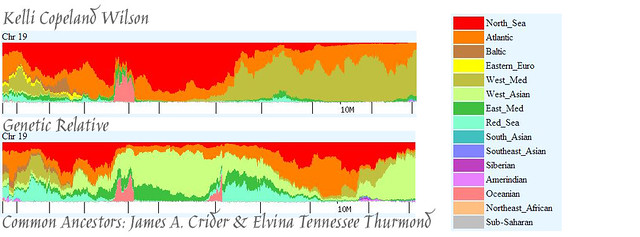
And, now, Chromosome 19. Here’s what Eurogenes K9 gave us. Yikes!
And, here is what happens when we use Eurogenes U15.
Oh, dear. This might have made things even more confusing. But, let’s give it a try. Whereas at first glance it appears that we match pretty much everywhere, there is something that gives us a clue. Bev’s segment has a rather thick stripe of West Asian running right through the middle of it. Mine does not. When looking at these segments it is helpful to remember that, given the genetic distance between me and Bev, we are actually looking at two very different lineages, with only a few bits that came from the same person. In people with predominantly European DNA, one would expect to see patterns repeat themselves without necessarily indicating a close relationship. So, whereas the top portion of our segments match, that is not entirely surprising given that we are both fairly Western European (red and orange). What I suspect is that we actually match on the bottom portion of the segment, and we can use Bev’s West Asian DNA as a dividing line (including it with her European DNA at the top), since I don’t have that. We are left with a mix of green (Eastern Mediterranean) light blue (Red Sea), a spike of pink (Oceanian), and some little bits of dark purple (Siberian) and light purple (Amerindian).
Again, I wonder if this might be Elvina. It could also be the someone who lived a long, long time ago, maybe even thousands of years ago. A child gets 50% of his DNA from his mother, and 50% from his father. Anything farther back than that isn’t guaranteed to follow the same formula (meaning the child might not get 25% from each grandparent, 12.5% from each great-grandparent, etc) — it’s not a mathematically perfect transmission. What’s really interesting is that, while there is a 50% chance that a segment of DNA will be passed on, if it is passed on, there is a 90% chance that it will stay largely intact (see Sources at bottom of page). So, it’s possible for relatively large segments of DNA to be passed from generation to generation for hundreds, maybe even thousands, of years without diluting down to nothing.
The developer of the EU15 (and also the K9) utility suggests that we ignore anything that comprises less than 1% of our DNA. By that token, these little bits of Bev’s and my DNA would be irrelevant. But, are they, really? It’s clear that Bev and I share these segments, as elucidated above, so am I supposed to believe that these seemingly anomalous bits of DNA are, in fact, nothing? Isn’t it also possible that they could be the genetic signature of an ancestor that lived so long ago that we’ll never be able to identify him or her? Because of the way Bev’s and my DNA is nestled together, we can imagine those stripes and spikes of color as a way of looking back through time: the green (Eastern Mediterranean) is most recent, then father back is the light blue (Red Sea), and then all the way back in the beginning is the pink (Oceanian). This person’s ancestors migrated from somewhere in the Polynesia area, to the Middle East, and then settled somewhere along the Eastern coast of the Mediterranean. And, that, cousins, was undoubtedly a long, long time ago. At some point, the descendant of this ancient lineage encountered an individual who was carrying Native American DNA, as well (which we can see, in a very small amount, in the parts we share).
Do some geneticists (or software developers — let’s make sure we’re not pointing fingers in the wrong direction here) want us to deny the existence of these ancestors just because their appearance in our DNA is unexpected (or because the traditional types of genealogical evidence cannot “prove” that they belong in our family tree)? Even though I am just dipping my toes in the water of genetic genealogy I have the sense that, in the future, we will be rethinking our assumptions about minority admixture. I wonder if it’s possible that some of the pieces of DNA that we see in our genome today are very, very old, and that the DNA from some of our more recent ancestors never made it into our genome at all. Dismissing small pieces of anomalous DNA as “noise” is a denial of what our genes are trying to tell us — the mysteries of the past persist.
Your cousin,
Sources:
Estes, Roberta. “Why Are My Predicted Cousin Relationships Wrong?” DNAeXplained, October 21, 2013, http://dna-explained.com/2013/10/21/why-are-my-predicted-cousin-relationships-wrong/